Jian Zhang, Ph.D.
Associate professor
Academy of Medical Engineering and Translational Medicine, Tianjin University
Room 107, Office Building, AMT, Tianjin University, No. 92, Weijin Road, Nankai District, Tianjin, Chinajian_zhang@tju.edu.cn
Google Scholar
BIOGRAPHY
Dr. Zhang obtained his bachelor‘s degree of computer science and technology at Nankai University in 2013. He got his Ph.D. in bioinformatics at Beijing Institute of Basic Medical Sciences, Beijing, China in 2018, under the supervision of Dr. Xiaomin Ying. From 2016 to 2018, as a Ph.D. visiting scholar, he worked with Dr. Xiaole Shirley Liu at Dana-Farber Cancer Institute and Harvard University. In 2020, Dr. Zhang joined Tianjin University as the Peiyang Young Scientist to continue his research on computational biology and bioinformatics. His group focus on developing bioinformatics methods and machine learning technologies for investigating high-throughput omics data to promote accurate diagnosis and personalized treatment of cancer and many other diseases.
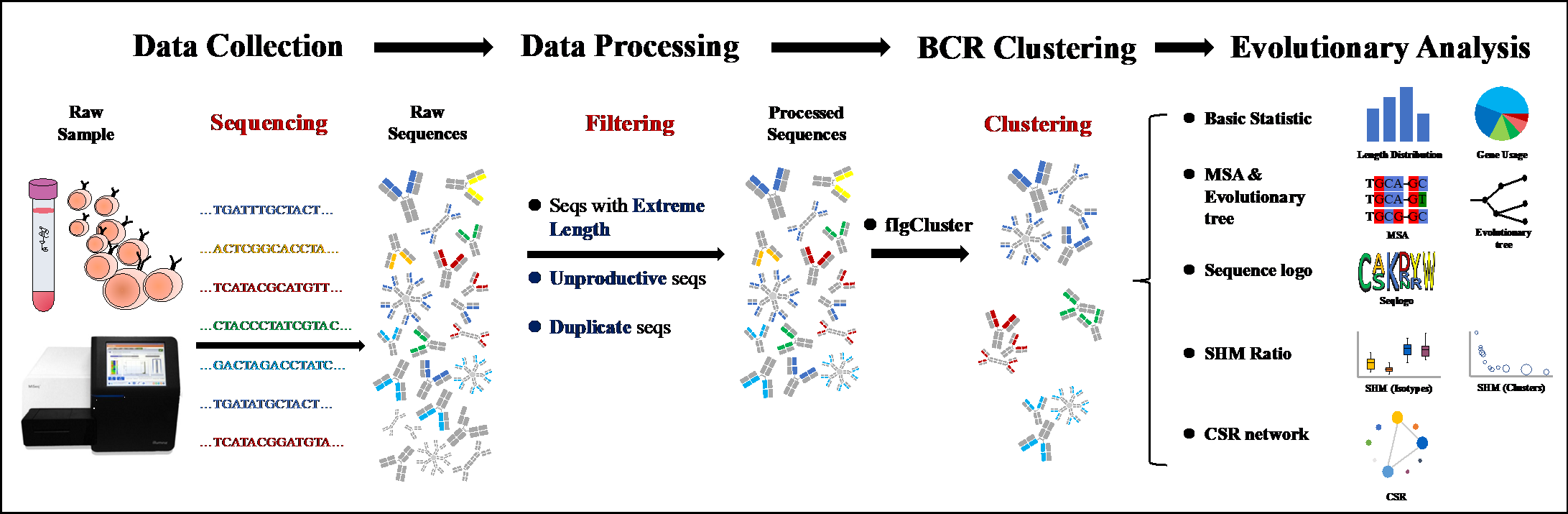
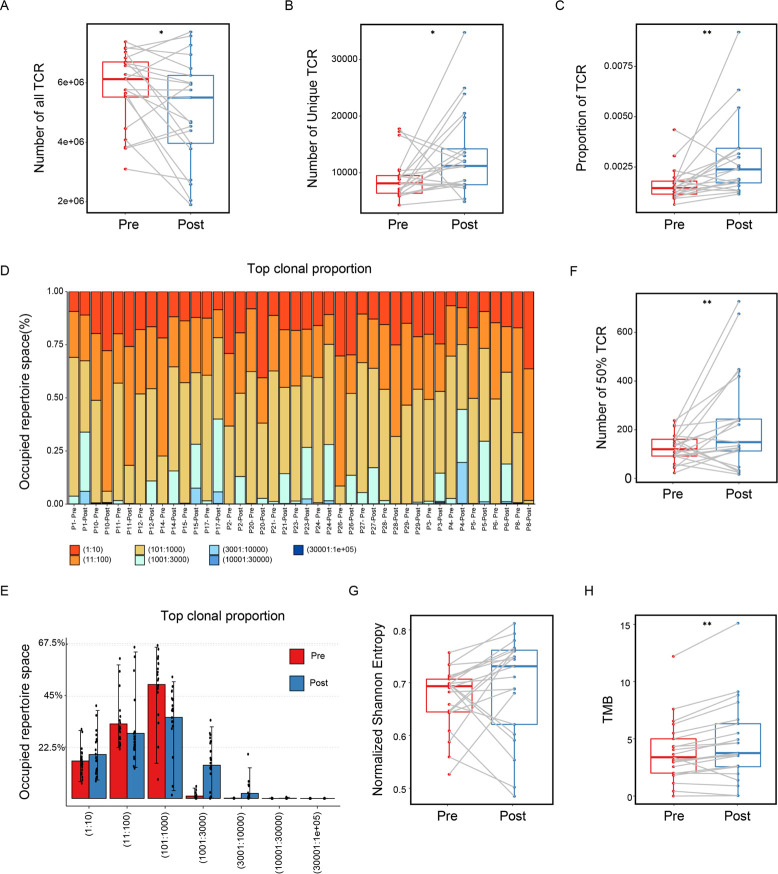
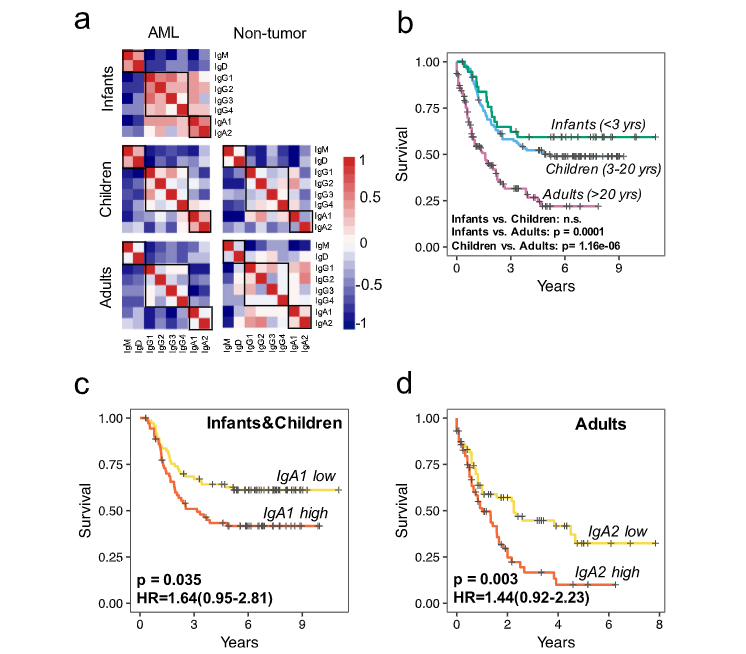
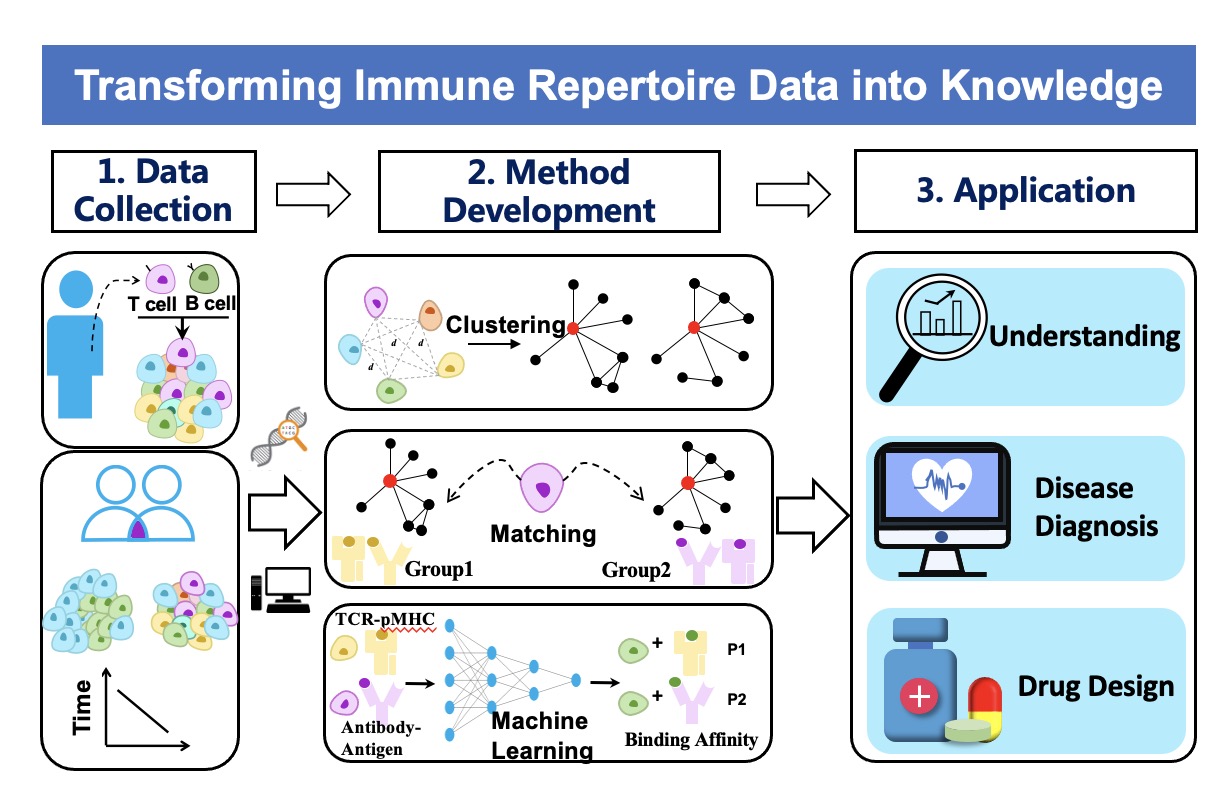 T and B cells are the two cores of the adaptive immune system, and both express antigen-specific receptors at their surface, namely, T cell receptors (TCRs) and B cell receptors (BCRs). The broad diversity of TCR and BCR ensures that the adaptive immune system is capable of recognizing a vast array of antigens, including pathogens, auto-antigens, allergens, and tumor neoantigens. Thus profiling and characterizing those antigen-specific receptors has become an important part of basic and clinical immunology research. We are interested in developing computational technologies to investigate the link between TCR/BCR sequence and epitope binding specificity, and explore their potential for disease detection, drug design and monitoring the responses to therapy. We especially hope that our research work can further deepen the understanding of our immune system.
T and B cells are the two cores of the adaptive immune system, and both express antigen-specific receptors at their surface, namely, T cell receptors (TCRs) and B cell receptors (BCRs). The broad diversity of TCR and BCR ensures that the adaptive immune system is capable of recognizing a vast array of antigens, including pathogens, auto-antigens, allergens, and tumor neoantigens. Thus profiling and characterizing those antigen-specific receptors has become an important part of basic and clinical immunology research. We are interested in developing computational technologies to investigate the link between TCR/BCR sequence and epitope binding specificity, and explore their potential for disease detection, drug design and monitoring the responses to therapy. We especially hope that our research work can further deepen the understanding of our immune system.
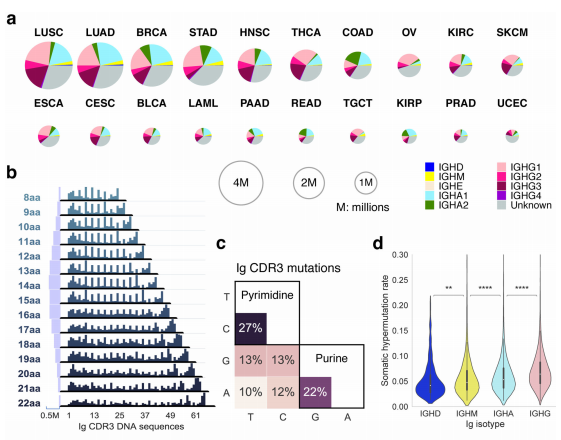
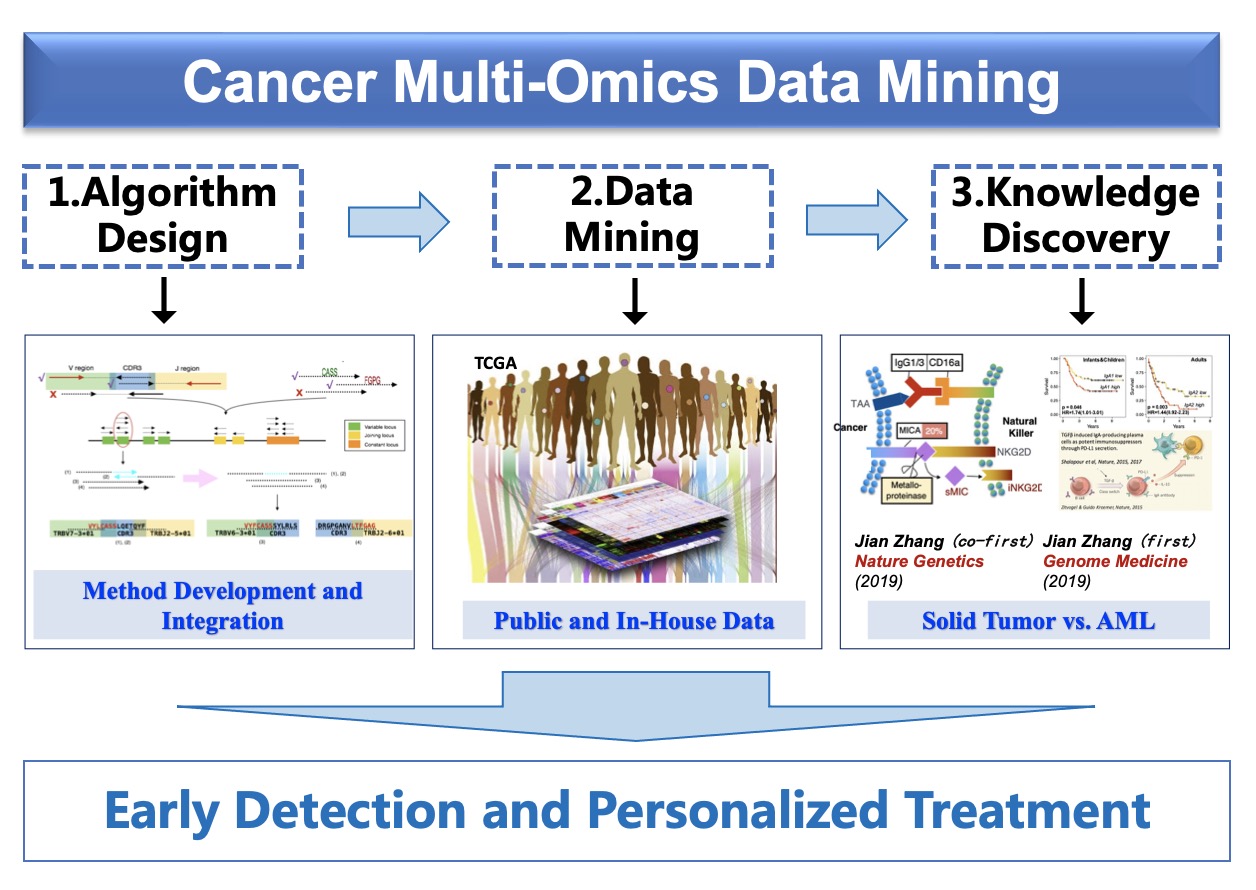 The advent of omics technologies, such as genomics, epigenomics, transcriptomics and proteomics, have revolutionized medical research. The concept of “omics” refers to the fact that all or nearly all instances of the targeted molecular space are measured in the assay, and therefore they provide holistic views of the biological system. The efficient processing and analysis of the omics data, especially extracting and discovering patterns from these data, has brought many challenges to bioinformatics and computational biology.We are interested in developing computational methods to mine large-scale omics data to improve understanding of the causes and biological processes of diseases, especially in cancer, and to identify signatures and build models for early cancer detection and personalized treatment.
The advent of omics technologies, such as genomics, epigenomics, transcriptomics and proteomics, have revolutionized medical research. The concept of “omics” refers to the fact that all or nearly all instances of the targeted molecular space are measured in the assay, and therefore they provide holistic views of the biological system. The efficient processing and analysis of the omics data, especially extracting and discovering patterns from these data, has brought many challenges to bioinformatics and computational biology.We are interested in developing computational methods to mine large-scale omics data to improve understanding of the causes and biological processes of diseases, especially in cancer, and to identify signatures and build models for early cancer detection and personalized treatment.